 Over the past several years, studies have revealed an astonishing diversity in our so-called microbiome. A five year project utilising researchers from around the world has been constructed to identify our mutual cohabitants that define our microbiome.[1] In Europe the MetaHIT project has pulled 8 countries and 13 academic partners together to add further data to this project.[2]
Over the past several years, studies have revealed an astonishing diversity in our so-called microbiome. A five year project utilising researchers from around the world has been constructed to identify our mutual cohabitants that define our microbiome.[1] In Europe the MetaHIT project has pulled 8 countries and 13 academic partners together to add further data to this project.[2]
If quantity matters, then the numerical assessment of the microbiota ensure that it won’t be found wanting. The gastrointestinal tract alone is inhabited with 1013–1014 microorganisms, a figure thought to be 10 times that of the number of human cells in our bodies and 150 times as many genes as our genome.[3] The estimated species number varies greatly but it is generally accepted that the adult microbiome consists of greater than 1000 species possibly as many as 40,000 [4] and more than 7000 strains.[5] It is a highly complex environment dominated by bacteria, mainly strict anaerobes, but also includes viruses, protozoa, archae, and fungi.[6] The microbiome is largely defined by two bacterial phylotypes, Bacteroidetes and Firmicutes with Proteobacteria, Actinobacteria, Fusobacteria, and Verrucomicrobia phyla present in relatively low abundance.[7]
The communities of resident microorganisms vary from individual to individual—even twin to twin—and from body part to body part. Researchers have been challenged many times over whether they would ever make sense of the composition of these unique communities or even be able to show how they affect human health. Interestingly most of the older medical philosophies have embraced the philosophy of treating the human body as a whole, complete system that needs to be balanced, which increasingly is understood to match up well with the synergistic properties of the gut microbiome. Of particular note for people employing the use of herbal products, Prof Nicholson at Imperial College London has demonstrated (unpublished) that after taking Chamomile that there are changes in the gut bacteria that are still present months after discontinuation suggesting that this and other herbs causes a lasting shift in gut microbe metabolism. Other work indicates that microbiota manipulation is a useful tool for beneficially modulating xenobiotic metabolism and pharmacokinetics in personalised health care.[8]
In 2011, researchers described what they felt was a recognisable pattern amid the complexity. A European consortium evaluated the gut microbial makeup of 22 Europeans using differences in a bacterial gene to distinguish the species within and between individuals. They compared these microbiomes with about a dozen previously characterised in Japan and the United States.
Far from being random, it turns out that our internal microbial communities fell roughly into three enterotypes, which the researchers dubbed Bacteroides, Prevotella, and Ruminococcus after the dominant microbe in each.[9] The gut microbiomes from larger samples of 154 Americans and 85 Danes also fitted well into three groups, indicating that there are a limited number of well-balanced communities in the human gut. The classifications however, were not correlated with people’s age, weight, sex, or nationality. Each enterotype differed in how it processed energy and in which vitamins it produced, factors that could influence the health of the human host and demand different approaches to health restoration.[10]
More work is needed to confirm that enterotypes are real and if they actually posess any clinical relevance. However if you would like to clarify your own microbiome, or add to the research the scientists are offering a chance to have a stool sample fully analysed on their web site.[11] Meanwhile, another team found that types seem to correlate with diet.[12] For example, the organism Bacteroides thrives on high-meat diets; Prevotella seems to favour vegetarian fare. Yet both of these organisms have a stubborn resistance to quick change as demonstrated when neither enterotype was affected by 10 days of dietary restrictions, suggesting that they are more influenced by long-term eating trends, albeit that other studies suggest that some species are quick to respond to dietary changes.[13]
This year, researchers also made other strides in understanding how diet affects the microbiome. They introduced 10 human gut bacteria into germ-free mice and monitored the composition of the bacterial community as the mice consumed different proportions of protein, fat, starch, and sugar. The results suggested rules for predicting how a change in food will alter the abundance of each species. The approach will help clarify the interplay between diet and microbes in nutrition and disease.[14],[15]
Several other studies provided more clues about the microbiome’s role in disease, development, and immune function.[16] Delving into the microscopic components found inside and on our microbiome, researchers continued to characterise the virome: all the viruses of the body. Far from being alien invaders, our microbes are integral to who and what we are and their role in human health as well as disease presents an even more complicated picture.[17],[18]
Comment
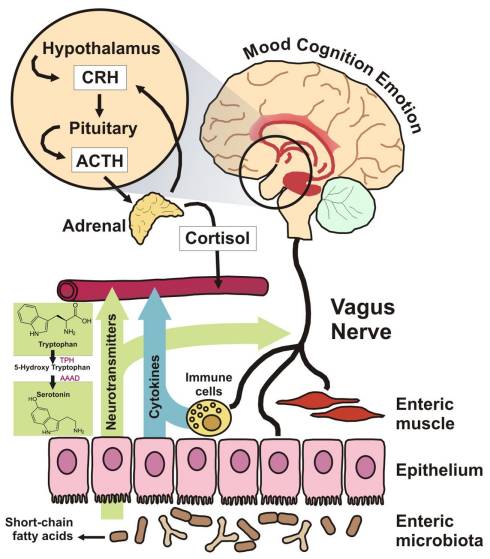
These are just a few of the many papers that have emerged in the scientific and clinical journals over the last 12 months and hint at many exciting discoveries yet to come.
When I began writing and clinically utilising the microbiome to manipulate the adverse effects of disordered mucosal immune systems back in the 1980’s I was regarded as a somewhat strange and very alternative practitioner. Much of my early work was on the role of the microbiome and the potential for manipulating the CNS and mood. It would be churlish to say this is a field of clinical care that is understood or even regularly successful, but as species and pathways are clarified, it represents a great opportunity for specific food choices and bacterial supplementation in the years to come, that may alleviate much of the CNS driven ailments of western society.
Therefore I shall finish with one reference: ‘Brain–Gut–Microbe Communication in Health and Disease’, published in 2011, and available a free full paper that I recommend anyone interested in this field to read and enjoy the evolutionary changes this approach is unveiling.[19] The image to the right is extracted from this paper.
References
[1] Microbiome Project. https://commonfund.nih.gov/hmp/
[2] Meta HIT Project: http://www.metahit.eu/index.php?id=410
[3] Qin J, Li R, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010 Mar 4;464(7285):59-65. View Abstract
[4] Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era.Curr Opin Gastroenterol. 2008 Jan;24(1):4-10. View Abstract
[5] Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006 Feb 24;124(4):837-48. Review. View Abstract
[6] Xu J, et al Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007 Jul;5(7):e156. Epub 2007 Jun 19. View Abstract
[7] Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005 Jun 10;308(5728):1635-8. Epub 2005 Apr 14. View Abstract
[8] Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want EJ, de Waziers I, Cloarec O, Richards SE, Wang Y, Dumas ME, Ross A, Rezzi S, Kochhar S, Van Bladeren P, Lindon JC, Holmes E, Nicholson JK. Colonization-induced host-gut microbial metabolic interaction. MBio. 2011 Mar 1;2(2):e00271-10. doi: 10.1128/mBio.00271-10. Print 2011. View Full Article
[9] M. Arumugam et al., Enterotypes of the human gut microbiome, Nature 473, 174–180 (2011) View Abstract
[10] U. Gophna, The Guts of Dietary Habits, Science 334, 45-46 (2011). View Abstract
[11] Bacterial Analsis: http://my.microbes.eu/index.cgi
[12] G. D. Wu et al., Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes, Science 334, 105-108 (2011). View Abstract
[13] Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011 Jan;12(1):5-9. View Abstract
[14] J. J. Faith et al., Predicting a Human Gut Microbiota’s Response to Diet in Gnotobiotic Mice, Science 333, 101-104 (2011). View Abstract
[15] B. D. Mueggeet al., Diet Drives Convergence in Gut Microbiome Functions Across Mammalian Phylogeny and Within Humans, Science 332, 970-974 (2011). View Abstract
[16] A. L. Kau et al., Human nutrition, the gut microbiome and the immune system, Nature 474, 327–336 (2011). View Abstract
[17] E. Pennisi, Gut Bacteria Lend a Molecular Hand to Viruses, Science 334, 168 (2011). View Abstract
[18] E. Pennisi, Going Viral: Exploring the Role Of Viruses in Our Bodies, Science 331, 1513 (2011). View Abstract
[19] Grenham S, Clarke G, Cryan JF, Dinan TG. Brain-gut-microbe communication in health and disease. Front Physiol. 2011;2:94. Epub 2011 Dec 7. View Full Paper





